First off, I recommend always setting your working directory. This means that you set up to which folder all data is coming in and out of and that way you won't mess up with typing in the directories over and over again. You do it this way:
setwd("C:/Projects/Rforpublichealth")
Great, now that that is done, you can start importing and exporting:
(1) Excel files
This is the easiest one - R has built in functions called read.csv() and write.csv() that work great. They maintain first line variable headings. I find that if you have an .xls file, the best thing to do is to save it as a .csv before trying to input into R. There are ways to get .xls into R but it's not worth the trouble since converting to a .csv takes 5 seconds. The function looks like this:
mydata<-read.csv(file="mydata.csv")
There are options for not maintaing first line variable headings (Header=FALSE), but I can't really think of a time that you would want to do that. You can also change what symbol is delimiting your data (tab delimited for example with sep = "\t") , but again I have never used those options. If you're in need of those options, you can read about them here.
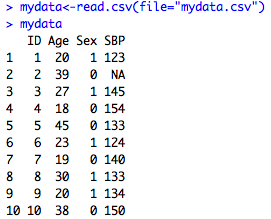
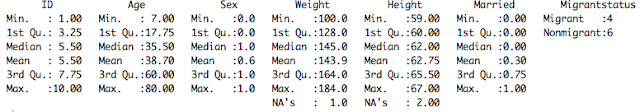
As you see on the left, R will automatically convert missing cells to NA, meaning that it's a numeric variable. If you have periods or something else that indicates missing in your CSV file, you could do a find and replace of the csv file for periods and change them to blanks.
Otherwise you can fix it with an ifelse() statement like:
mydata$SBP<-as.numeric(ifelse(mydata$SBP==".", NA, mydata$SBP))
Or with an apply() statement. Check out my earlier blog post about this.
Back to importing: If you get an error like the one you see to the right, 99.9% of the time you have misspelled the name of the file or put it into the wrong folder. Always check to make sure the directory name and the file name match up and all your slashes are in the right place.
Now, let's say you change some things and want to export your data back into a csv. To export, you do:
write.csv(mydata, file="mynewcsvfile.csv")
(2) Stata .dta files
For this, you will need the foreign package. You must first install the foreign package (you just use the menu bar in R, find the "Install packages" option and go from there). Then you must then load the library this way:
library(foreign)
Now you can use the read.dta() and write.dta() functions. They are very similar to the read and write csv functions, and look like this:
mystatadata<-read.dta(file="mystatafile.dta")
write.dta(mystatadata, file="mystatafile2.dta")
(3) SAS files
(a) SAS transport files
SAS can be tricky because it uses both transport (XPORT) files as well as sas7bdat files and they are very different in terms of their encoding. The XPORT files are most common and easier to work with. For these, you need to first install and load the SASxport library
library(SASxport)
Now, you can use the read.xport() function in the same way we have used the other read functions:
mysasdata<-read.xport(file="mysasfile.xpt")
And again we can export:
write.xport(mydata, file="mynewsasfile.xpt")
(b) SAS7bdat files
This used to be a huge pain. Fortunately, there is a new package for reading these files into R called the sas7bat package. First, install this package and load. Then use the read.sas7bdat() function:
library(sas7bdat)
mysas7bdat<-read.sas7bdat(file="myothersasfile.sas7bdat")
However, I have personally had some problems with this function not working for me. Hopefully it will work for you. If it doesn't, you can try to convert the SAS file using StatTransfer or with JMP if you don't have SAS on your computer.